Introduction
MicroRNA (miRNA) is a type of endogenous non-coding RNA (ncRNA) with a regulatory function found in eukaryotes, typically ranging from 18 to 25 nucleotides in length. Mature miRNA is generated through a series of nuclease cleavages and processing of longer precursor transcripts. It is then incorporated into RNA-induced silencing complexes. These complexes recognize target mRNA through base-pair complementarity, guiding the RNA-induced silencing complexes to either degrade the target mRNA or inhibit its translation, depending on the degree of complementarity [1,2]. Recent studies have shown that miRNAs play roles in a variety of regulatory pathways. These include development, viral defense, hematopoiesis, organogenesis, cell proliferation and apoptosis, as well as lipid metabolism [2-5].
Exosomes have recently become a prominent topic, and the analysis of their cargo has garnered significant attention [6-9]. The double-layered lipid membrane of exosomes ensures the stability of their internal contents, providing a more accurate reflection of the physiological and pathological states of their cells of origin. Consequently, exosome research has been incorporated into the realm of liquid biopsy, complementing studies on circulating tumor cells and cell-free nucleic acids [10]. Our previous research indicated that the detection of exosome-derived miRNAs is more consistent than that of mRNAs [11]. However, several questions remain unanswered: Are exosomal miRNAs uniformly expressed in different bodily fluids, such as blood and urine, within the same human body? Which exosomal miRNA is present across various bodily fluids under normal physiological conditions? Which body fluid is more suitable for the detection of extracellular vesicles in future studies? Currently, these issues remain unresolved. This study presents a comparative analysis of exosomal miRNAs in urine and blood under physiological conditions.
Methods
This study was approved by the Ethics Committee of Chengdu University (no. 2081920062). All participants understood the purpose and content of the study and signed informed consent.
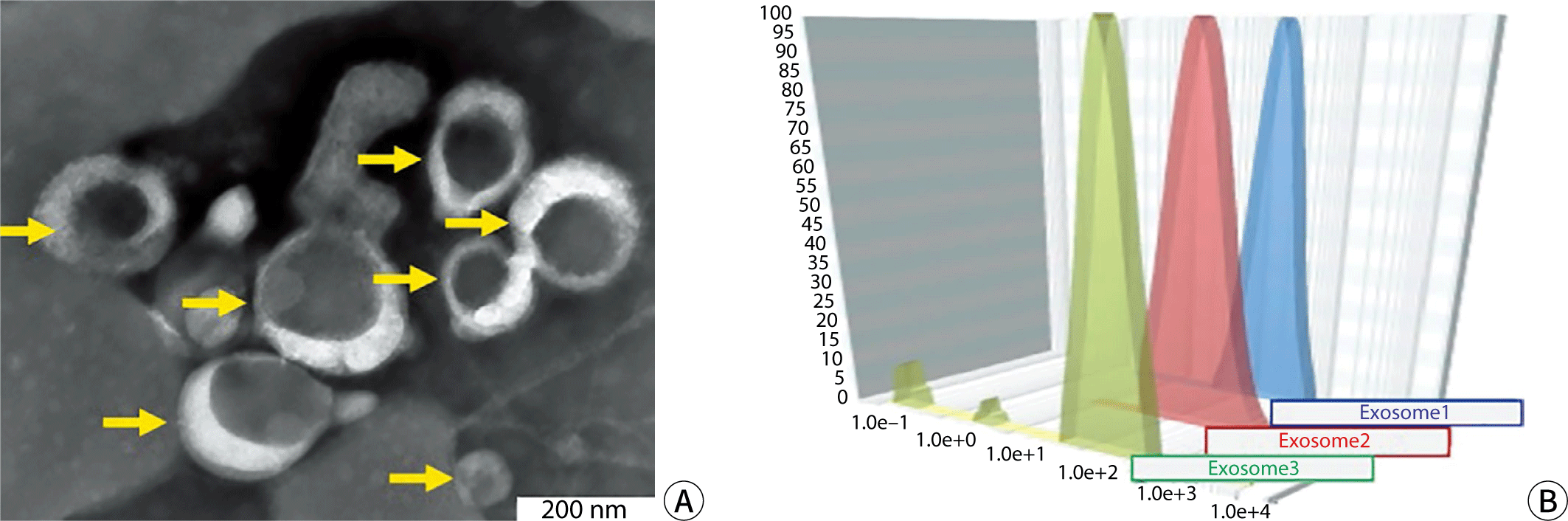
Urine samples were obtained from morning midstream collections, and blood samples were drawn from fasting individuals who had not eaten for more than 10 hours—specifically, 10 mL of peripheral venous blood and 100 mL of urine were collected from six healthy adults. These volunteers had not taken any medications for at least seven days prior to sample collection. The basic information for the six individuals is presented in Table 1. Blood samples, treated with EDTA as an anticoagulant, were centrifuged at 1,000×g for 15 minutes, and the resulting supernatant was retained. After further centrifugation at 10,000×g for 10 minutes at 4℃, ammonium sulfate powder was added to achieve a final concentration of 1.06%. The mixture was then centrifuged again at 10,000×g for 15 minutes at 4℃, and the supernatant was aspirated. An equal volume of 16% polyethylene glycol solution was added to the remaining solution and left to stand overnight at 4℃. Following centrifugation at 4,000×g for 1 hour, the supernatant was carefully removed, and the pellet, containing the exosomes, was collected. The enrichment and identification of urinary exosomes were carried out in accordance with established protocols [12,13]. The results of the electron microscopy examination of the exosomes and the particle size analysis are depicted in Fig. 1.
The sequencing was conducted in accordance with the manual provided by Illumina, which included steps for library preparation and sequencing experiments. The small RNA sequencing library was prepared using TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, USA). Once the library preparation was complete, the constructed library was sequenced on an Illumina HiSeq 2000/2500 platform, utilizing a single-end read length of 50 bp.
FastQC version 0.10.1 was utilized for data quality control, ACGT101-miR version 4.2 for basic miRNA analysis, ACGTUNAfold version 3.7 for advanced miRNA analysis, TargetScan version 5.0 and Miranda version 3.3a for miRNA target gene prediction, Pathway Network version 1.6 for target gene function analysis, and R software version 3.0.1 for mapping of result files. The databases used in the study are listed in Table 2.
| Database | Web address | Version date |
|---|---|---|
| miRNA (miRs) database | ftp://miRBase.org/pub/miRBase/CURRENT/ | 22.0 |
| Pre-miRNA (mirs/MIRs) database | ftp://miRBase.org/pub/miRBase/CURRENT/ | 22.0 |
| RFam | Collection of many common non-coding RNA families other than miRNA http://rfam.janelia.org | 12.0 |
| Repbase | Prototypic sequences representing repetitive DNA from different eukaryotic species http://www.girinst.org/repbase | 22.07 |
| Genome database | ftp://ftp.ensembl.org/pub/release-101/fasta/homo_sapiens/dna/ | |
| mRNA database | ftp://ftp.ensembl.org/pub/release-101/fasta/homo_sapiens/dna/ | |
| KEGG pathway database | http://www.genome.jp/kegg | 2016.05 |
| Gene Ontology database | http://geneontology.org/ | 2016.04 |
The experimental data are expressed as the mean±SE. The input data for miRNA differential expression analysis consisted of normalized values. The P-value was calculated using a model based on the normal distribution. When biological replicates were present, the t-test was used to analyze differences between two groups of samples, and analysis of variance was employed for analyzing differences among multiple groups.
Results
The total number of reads for urinary exosomes ranged from 23,699,840 to 34,395,054, and the total number of bases ranged from 1,208,691,840 to 1,754,147,754, with averages of 27,871,527.67±5,722,347.37 and 1,421,447,911±291,839,716, respectively. The base composition ratios of adenine (A), thymine (T), cytosine (C), and guanine (G) varied from 21.94% to 31.81%. The Q20 and Q30 quality scores of the reads were between 96.14% and 99.29%, and the overall GC content of the reads ranged from 49.55% to 53.97%. For blood exosomes, the total number of reads and total bases were 17,002,521 to 38,704,493 and 867,128,571 to 1,973,929,143, respectively. The averages were 2,588,481.33±11,374,109.12 for reads and 1,320,128,948±580,079,565 for bases. The base composition ratios of A, T, C, and G were between 22.69% and 30.08%. The Q20 and Q30 quality scores of the total reads ranged from 91.61% to 99.40%, with the overall GC content of the reads between 52.55% and 52.91%. As indicated in Table 3, there was no significant difference between the groups (P>0.05).
Q20% reflects the quality of the data, indicating that in the sequencing results, the probability of a base error at a certain position caused by the sequencing instrument is less than 1%; Q30% indicates that in the sequencing results, the probability of a base error at a certain position caused by the sequencing instrument is less than 0.1%.
Rfam is a database of ncRNA families, which includes ribosomal RNA (rRNA), transfer RNA (tRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), miRNA, and other ncRNAs. The annotated sequences of rRNA, snoRNA, snRNA, tRNA, and other RNAs were extracted from the clean data produced by the sequencing machine, using the Rfam database as a reference. The results are presented in Table 4 and have been statistically analyzed and visualized, showing the mean of total reads and unique reads in Fig. 2. There were no significant differences observed between the groups (P>0.05).
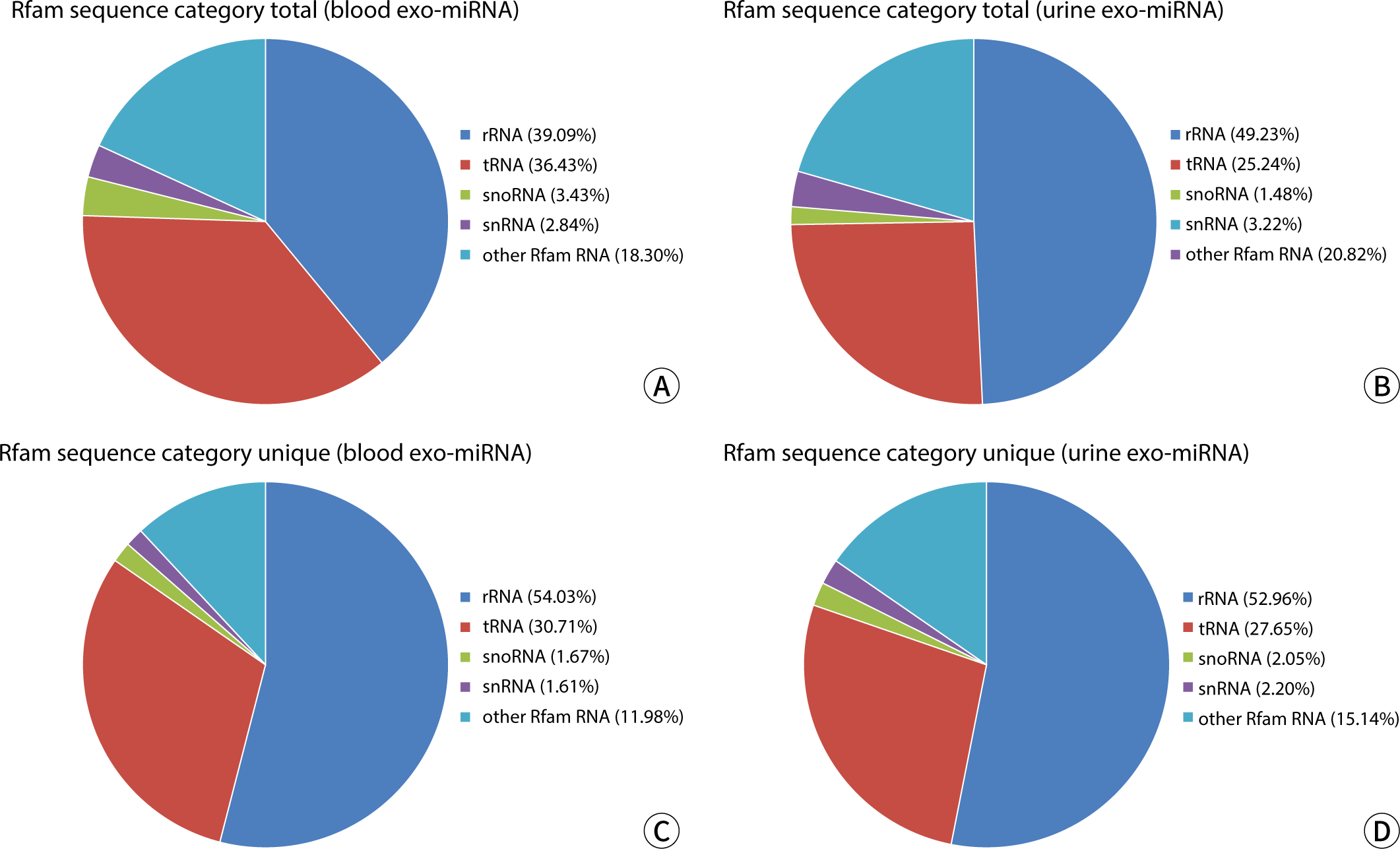
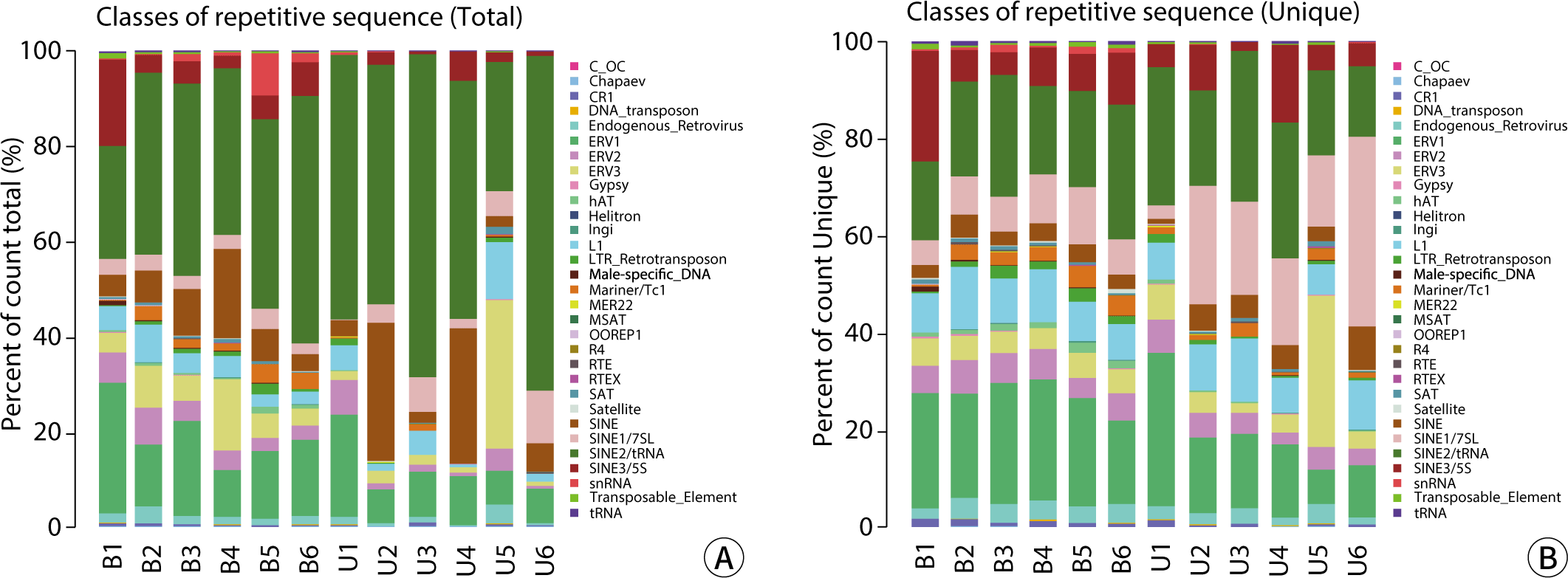
The Repbase database is a comprehensive collection of repeat sequences in eukaryotic DNA, encompassing transposable elements, tandem repeat sequences (such as satellite sequences or microsatellites), scattered genomic repeat sequences, and certain multicopy host genes (including rRNA, tRNA, and histone genes). Annotated repeat-associated sRNA sequences were extracted from the clean data produced by the sequencing machine, using the Repbase database as a reference. The data for total reads and unique reads were analyzed both statistically and visually (Fig. 3). The categorized data from the Repbase database are presented in Table 5. There was no significant difference between the blood and urine groups, with the exception of the total L1 sequence (P<0.05).
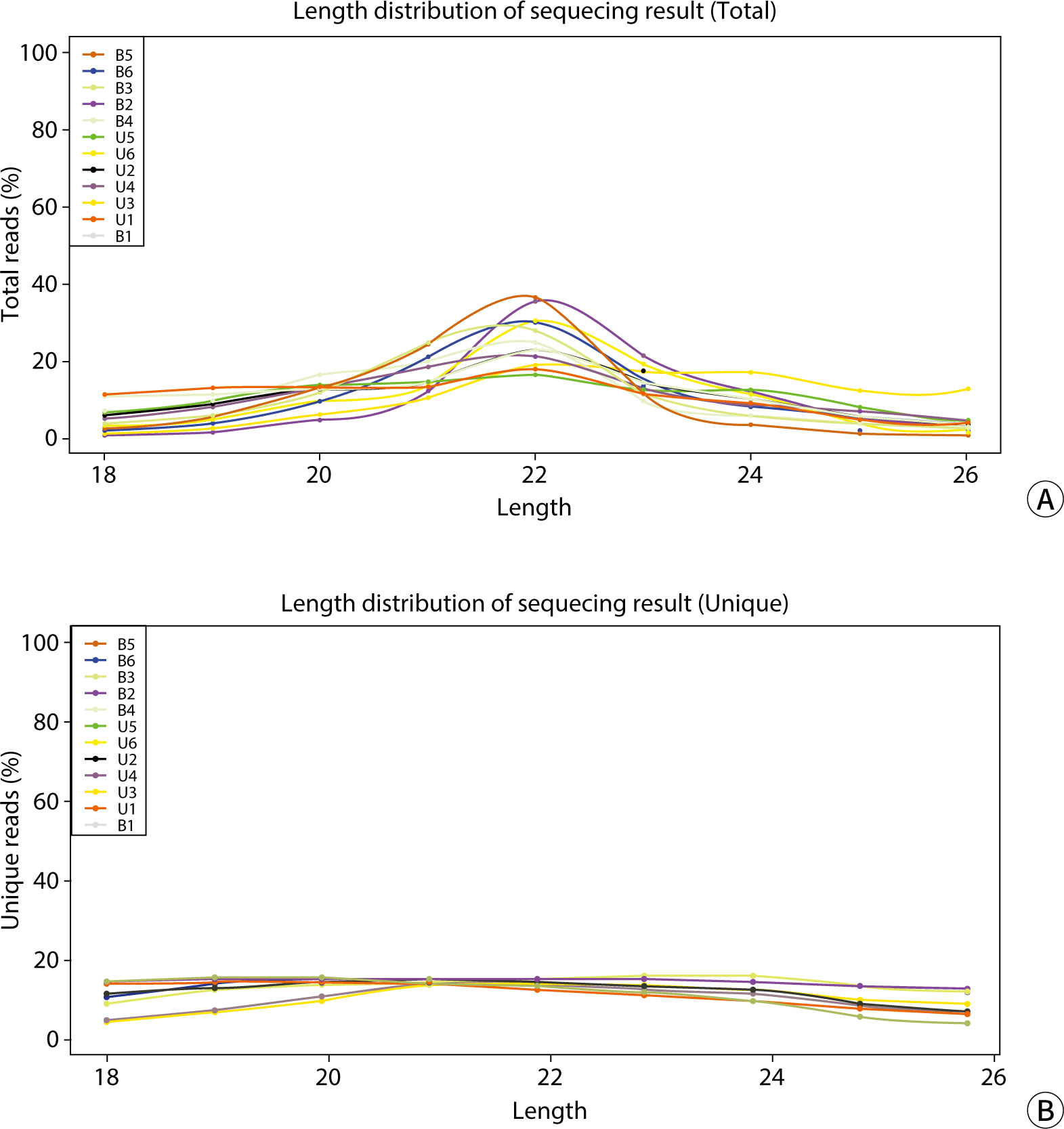
Based on the analysis and statistics of the original sequencing data, we carried out an assessment of the length distribution for both the total and unique numbers of the filtered valid data. As depicted in Fig. 4, the total reads were primarily concentrated in the 22-nucleotide length region. The unique reads were predominantly found within the 19–21 nucleotide range, with a slightly lower distribution at lengths 18 and 22–24, and minimal presence at length 26. When comparing the results from blood and urinary exosomes (Table 6), significant differences were observed between the groups in the 20–23 and 25–26 nucleotide lengths, as well as in the valid reads (P<0.05) for unique reads. However, no significant differences were found between blood and urine in total reads and other unique read lengths.
The accuracy of miRNA identification is highly correlated with the accuracy of miRBase, the genome of the species in question, and the completeness of the database. Clean data, which are essential for analysis, were utilized to identify small RNAs. The miRNA expression levels in each sample were determined using ACGT101 miR software. These expression levels were then used to evaluate the correlation of gene expression characteristics both within and between groups, as well as to evaluate the differential expression of miRNAs. When measuring expression levels, the normalized value (standardized based on the original miRNA read counts) was employed as the metric for miRNA expression, enabling the comparison of miRNA expression across different samples.
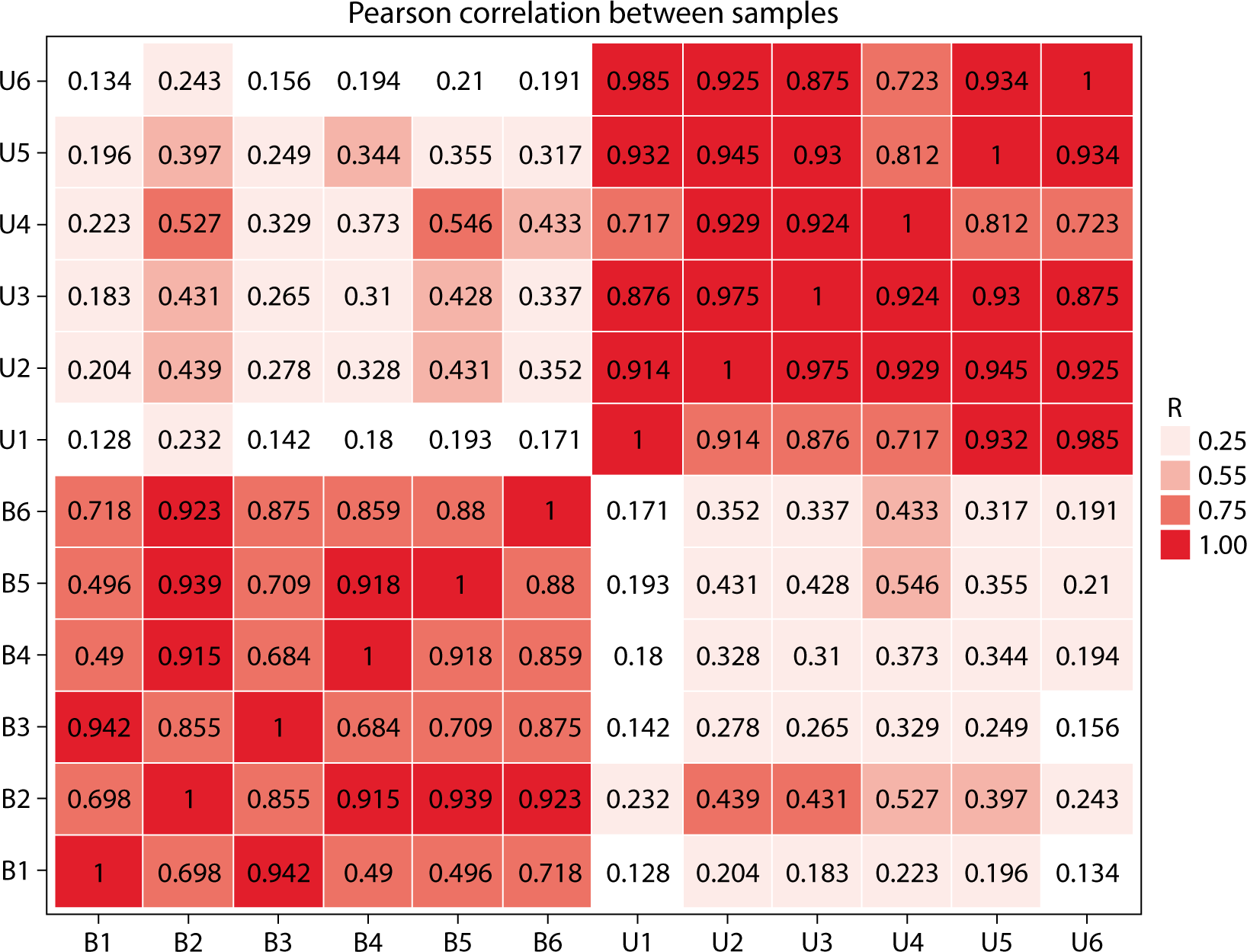
Correlation analysis, utilizing gene expression data from samples, can more accurately determine the clustering relationship among samples. A higher correlation coefficient indicates better clustering of the samples. By employing the Pearson correlation coefficient, we can assess the repeatability among samples and identify potential outliers. As shown in Fig. 4, the correlation coefficients between the six blood and urine samples ranged from 0.49 to 1. Specifically, blood samples had correlation coefficients ranging from 0.49 to 1, while urine samples had coefficients between 0.717 and 1. Notably, the correlation coefficients for the fourth urine sample and the first blood sample were relatively low when compared to the other samples (Fig. 5).
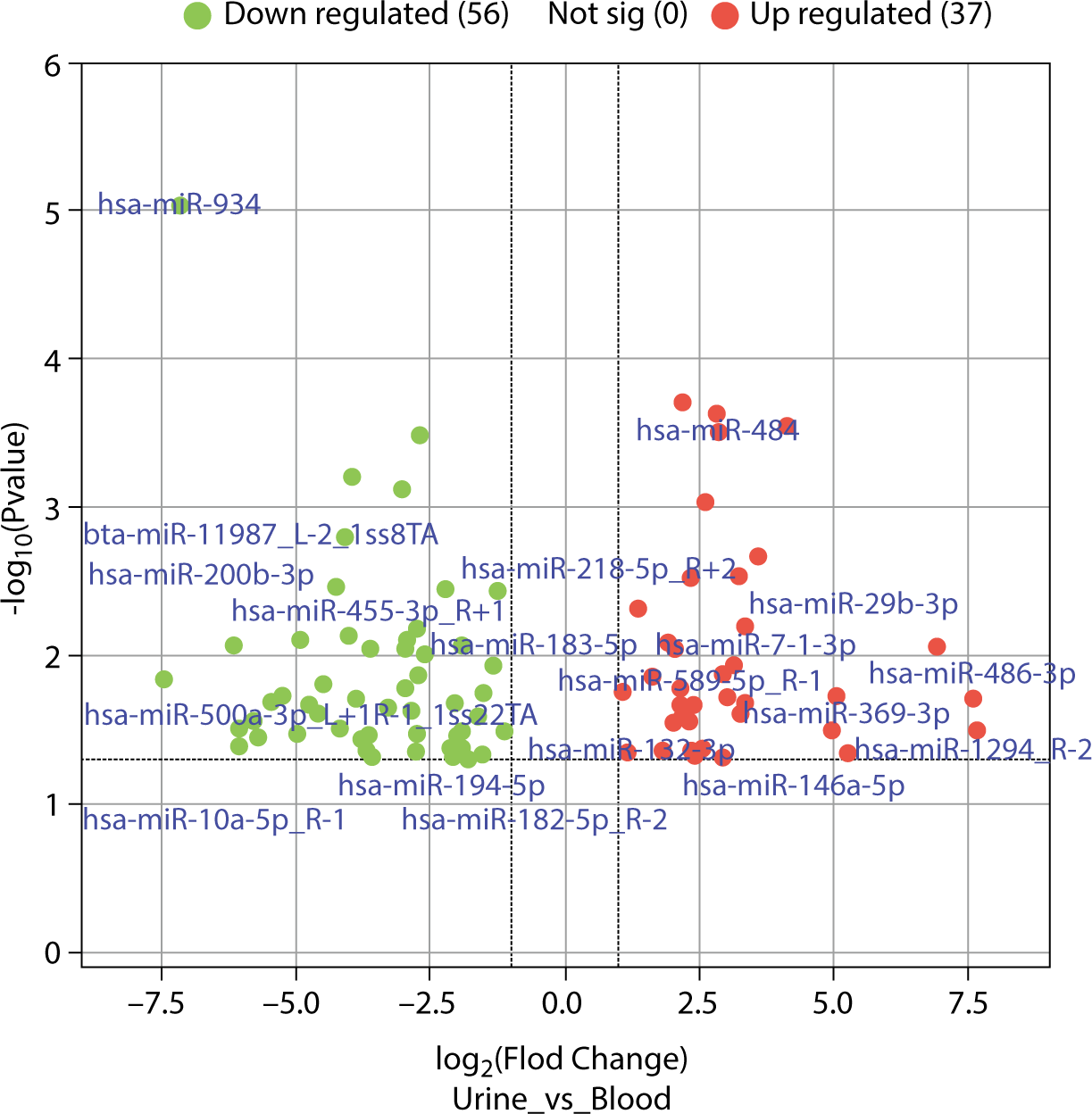
The differences in exosomal miRNAs between blood and urine were analyzed using the R language "limma" software package, identifying 102 differentially expressed miRNAs with fold changes greater than 2. Of these, 41 were more highly expressed in blood than in urine samples. The top 10 exosomal miRNAs with significant differences included hsa-miR-934, hsa-miR-30e-3p_1ss22CT, bta-miR-11987_L-1_1ss8TA, hsa-miR-10a-3p_R-1, bta-miR-11987_L-2_1ss8TA, hsa-miR-10b-5p_R-1, hsa-miR-218-5p_R+2, hsa-miR-30d-5p_R+2, hsa-miR-29c-5p_R-1, and hsa-miR-200b-3p. Conversely, 61 exosomal miRNAs were found at lower levels in blood than in urine samples, with the top 10 exosomal miRNAs showing significant differences being hsa-miR-425-5p, hsa-miR-7-5p_R-1, hsa-miR-145-5p, hsa-miR-484, hsa-miR-329-3p, hsa-miR-96-5p_R-2, hsa-miR-143-3p_R+1, hsa-miR-130b-5p_R+1, hsa-miR-140-3p_R+1, and hsa-miR-338-5p_R-1. The differentially expressed exosomal microRNAs (DEexo-miRNAs) are displayed in Fig. 6, excluding exosomal miRNAs with a log fold change (logFC) of Inf (4 instances) and -Inf (6 instances).
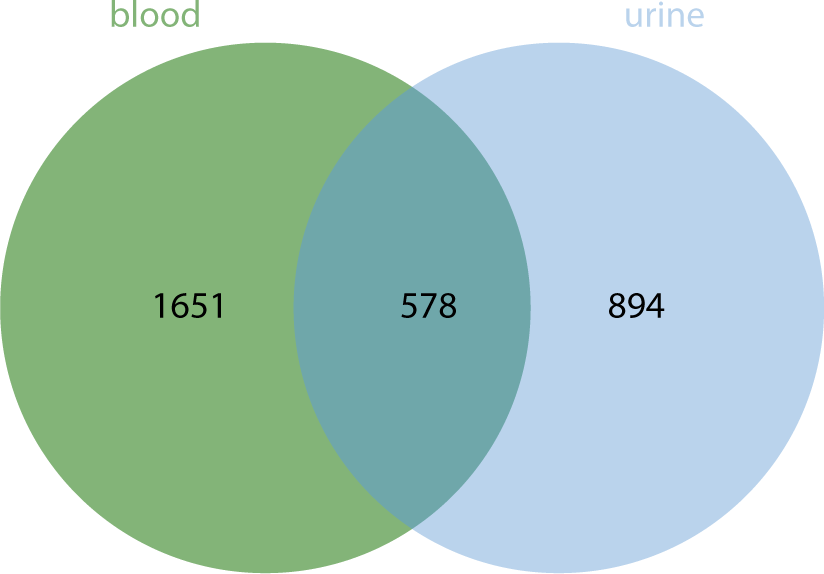
ENCORI (https://rnasysu.com/encori/) was utilized to predict the target genes (mRNA) corresponding to DEexo-miRNAs. Only those DEexo-miRNAs that matched targets in both TargetScan and Miranda were selected as the final target genes. For the top 10 DEexo-miRNAs in urine, 1,472 target mRNAs were identified, including DDX3Y, RERE, DFFA, RSRP1, PUM1, C1orf216, STK40, YRDC, RRAGC, RAB3B, CC2D1B, ZRANB2, EVI5, ARHGAP29, CLCC1, SORT1, SLC16A1, NRAS, SV2A, GATAD2B, and others. Similarly, for the top 10 DEexo-miRNAs in blood, 2,229 target mRNAs were obtained, including IFFO2, STMN1, FOXJ3, CTSS, RNF11, SRSF11, ATP1B1, LARP4B, AP3M1, TM9SF3, PTEN, LCOR, TIMM10, KMT5B, CREBZF, GNS, HSPB8, EFNB2, CFL2, and POMT2, among others. A total of 578 identical target mRNAs corresponding to DEexo-miRNAs were found between blood and urine (Fig. 7).
The target mRNAs corresponding to DEexo-miRNAs underwent Gene Ontology (GO) / Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotations. In other words, the target mRNA served as an intermediary to link exosomal miRNAs with their respective functions.
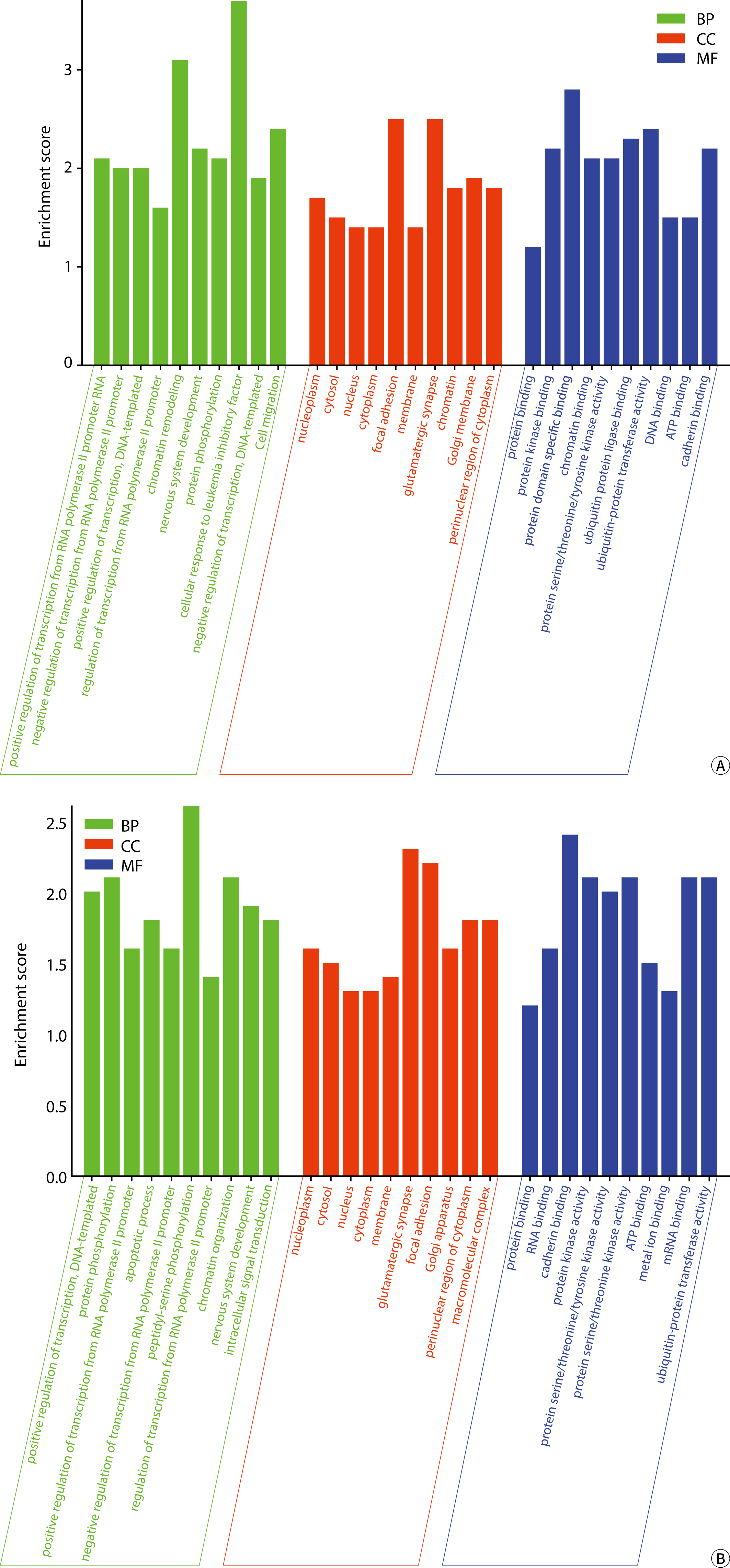
The GO annotation bar chart is shown in Fig. 8.
The biological processes (BPs) associated with the target mRNAs of exosomal miRNA in body fluids include positive and negative regulation of RNA polymerase II promoter transcription, DNA template/promoter activities, chromatin structure modulation, nervous system development, cell migration, and protein phosphorylation/polyubiquitination, among others. The BPs influenced by the target mRNA according to DEexo-miRNAs in urine also encompass chromatin remodeling, cellular response to leukemia inhibitory factor, ubiquitination of protein at the K48 linkage, cell adhesion via plasma membrane adhesion molecules (hemophilic interactions), proteasome-mediated ubiquitin-dependent protein catabolic processes, and peptide-threonine phosphorylation. The target mRNAs of DEexo-miRNAs in blood are involved in BPs such as the apoptosis process, peptide-serine phosphorylation, intracellular signal transduction, negative regulation of gene expression, post-embryonic development, cell cycle regulation, vesicle-mediated transport, the insulin receptor signaling pathway, and protein stability.
The cellular components (CCs) associated with the target mRNAs of exosomal miRNAs in two types of body fluids included the nucleoplasm, cytosol, nucleus, cytoplasm, adhesive spots, endoplasmic reticulum membrane, glutamatergic synapse, chromatin, Golgi apparatus membrane, cytoplasmic perinuclear area, macromolecular complexes, and the actin cytoskeleton, among others. In urine, the CC of the target mRNA encompassed cytoplasmic ribonucleoprotein granules/complexes, cell-cell junctions, cell projections, ubiquitin ligase complexes, and the cytoskeleton. The target mRNAs of DEexo-miRNAs in blood were associated with CC such as intracellular membrane-bounded organelles, nuclear specks, cytoplasmic stress granules, postsynaptic densities, early endosome membranes, and chromatin.
The molecular functions (MFs) associated with the target mRNAs of exosomal miRNA in two types of humor include protein (kinase/domain-specific) binding, protein serine/threonine/tyrosine kinase activity, ubiquitin protein ligase binding/transferase activity, ATP/cadherin/RNA/GDP binding, transcription factor activity with sequence-specific DNA binding, among others. The target mRNAs of DEexo-miRNAs in urine were involved in MFs such as chromatin/DNA/β-catenin/RNA polymerase II core promoter proximal region sequence-specific DNA binding, transcription coactivator/corepressor activity, transcription activator activity, RNA polymerase II transcription regulatory region sequence-specific binding, and protein kinase inhibitor activity. Meanwhile, the target mRNA in blood was associated with MFs including protein serine/threonine kinase activity, metal ion/mRNA/protein kinase/small GTPase binding, and bridging, and others.
KEGG enrichment analysis revealed that the signaling pathways targeted by exosomal miRNA-associated mRNAs in blood and urine are implicated in a variety of BPs and diseases. These include proteoglycans in cancer, axonal guidance, regulation of the actin cytoskeleton, cancer pathways, resistance to EGFR tyrosine kinase inhibitors, neurotrophic signaling pathways, endocytosis, cellular adhesion sites, cellular senescence, autophagy in animals, and Salmonella infection, among others. In urine, the targeted mRNAs are involved in pathways such as ubiquitin-mediated proteolysis, the phosphatidylinositol signaling system, the ErbB/Wnt signaling pathway, protein processing in the endoplasmic reticulum, endocrine resistance, gap junction communication, oocyte meiotic division, and Shigellosis. The mRNAs targeted by DEexo-miRNAs in blood are associated with the FoxO, MAPK, PI3K-Akt, Rap1, and Ras signaling pathways, as well as with chronic myeloid leukemia, bacterial invasion of epithelial cells, hepatitis B, and Yersinia infection. The KEGG enrichment histogram is presented in Fig. 9.
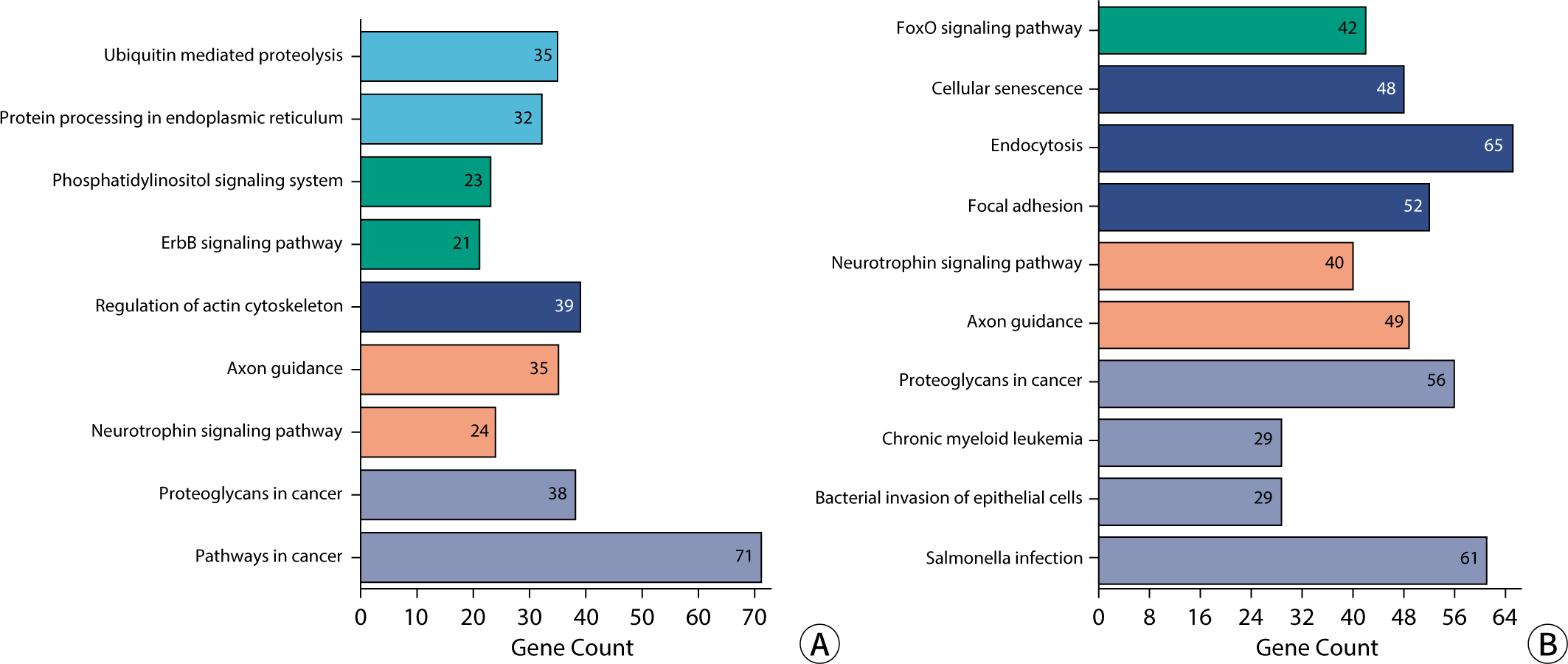
Discussion
Blood and urine samples are commonly used for clinical testing, as they contain a variety of substances that reflect the metabolic activity of tissues and organs within an organism. In other words, detecting the same substance in different specimens, such as blood or urine, can yield different results and indicate varying conditions of the body. Therefore, distinguishing the distribution of exosomal miRNAs in different body fluids is crucial for monitoring physiological states and establishing a basis for disease diagnosis. Some studies have reported on blood and urine miRNAs within the same article [14-16]. This paper presents the comparative results for the first time, and the biological analysis of these findings is also being reported for the first time.
The study revealed that the total read counts and the total number of bases in blood and urine samples were essentially identical, and the ratios of the A, T, G, and C bases were also remarkably similar. Comparative analysis of the Rfam database indicated that the content and proportion of rRNA, snoRNA, snRNA, tRNA, and other Rfam RNAs in blood and urine exosomes were fundamentally consistent.
The comparative analysis of the Repbase database revealed that the content and proportion of total reads and unique reads for various genetic elements—including CR1, DNA_transposon, Endogenous_Retrovirus, ERV1, ERV2, ERV3, Gypsy, hAT, Ingi, L1, LTR_Retrotransposon, Male-specific_DNA, Mariner/Tc1, OOREP1, MER22, MSAT, RTE, RTEX, R4, SAT, Satellite, SINE, SINE1/7SL, SINE2/tRNA, SINE3/5S, snRNA, Transposable_Element, and tRNA—in blood and urine exosomes were largely consistent. However, the total reads for L1 were higher in urine exosomes than in blood, although the unique reads showed no difference. The length distributions of candidate RNA exhibited variations between groups at lengths of 20-23 and 25, 26, as well as in the valid unique reads. There were no significant differences in total reads between blood and urine exosomes.
The analysis of DEexo-miRNAs revealed that 41 miRNAs were more highly expressed in blood samples than in urine, while 61 miRNAs showed lower levels in blood compared to urine. The top 10 exosomal miRNAs with higher expression in blood were hsa-miR-934, hsa-miR-30e-3p_1ss22CT, bta-miR-11987_L-1_1ss8TA, hsa-miR-10a-3p_R-1, bta-miR-11987_L-2_1ss8TA, hsa-miR-10b-5p_R-1, hsa-miR-218-5p_R+2, hsa-miR-30d-5p_R+2, hsa-miR-29c-5p_R-1, and hsa-miR-200b-3p. Conversely, the top 10 miRNAs with lower expression in blood included hsa-miR-425-5p, hsa-miR-7-5p_R-1, hsa-miR-145-5p, hsa-miR-484, hsa-miR-329-3p, hsa-miR-96-5p_R-2, hsa-miR-143-3p_R+1, hsa-miR-130b-5p_R+1, hsa-miR-140-3p_R+1, and hsa-miR-338-5p_R-1. These miRNAs correspond to 1,651 and 894 types of mRNA, respectively, with 578 types being common to both groups.
Studies have found that photosensitive aminotransferase 1 (PSAT1) is a target gene for hsa-miR-218-5p and hsa-miR-145-5p, which can be used to predict bone metastasis in gastric cancer [17]. Frørup et al. discovered that extracellular vesicles, particularly those enriched in the plasma exosomes of lactating mothers with Type 1 diabetes, contained abnormal levels of hsa-miR-30d-5p post-delivery [18]. Wu et al. identified plasma exosomal miR-103b, miR-877-5p, and miR-29c-5p as diagnostic biomarkers for early lung adenocarcinoma by combining bioinformatics with experimental verification [19]. Chen et al. reported the upregulation of urinary exosomal hsa-microRNA-200b-3p and hsa-microRNA-206 in patients with steroid-induced osteonecrosis of the femoral head [20].
Sinha et al. found that hsa-miR-425-5p was significantly upregulated, while hsa-miR-663a was downregulated under diabetic conditions [21]. Deng et al. discovered that exosomes derived from the plasma of septic patients inhibited apoptosis of T lymphocytes by downregulating Bcl-2-associated death promoter via hsa-miR-7-5p [22]. Zhang et al. found that hsa-miR-145-5p was significantly differentially expressed in the serum exosomes of patients with polycystic ovarian syndrome (PCOS) and considered these miRNAs as potential biomarkers for PCOS [23]. Hang et al. argued that the downregulation of miR-145-5p in cancer cells and their derived exosomes may contribute to the development of ovarian cancer by targeting connective tissue growth factor [24]. Zhao et al. revealed that the injection of RGD-modified exosomes loaded with miR-484 induced vessel normalization, which in turn sensitized cancer cells to chemotherapy-induced apoptosis [25]. Zhang et al. reported that low serum exosomal miR-484 expression predicted an unfavorable prognosis in ovarian cancer [26]. Li et al. identified cerebrospinal fluid exosomal hsa-miR-96-5p as a biomarker for diagnosing or monitoring the progression of non-small cell lung cancer with leptomeningeal metastases [27]. Tomita et al. considered urinary exosomal hsa-miR-143-3p as a predictive biomarker for persistent psychotic-like experiences [28]. Wang et al. found that hsa-miR-143-3p expression was significantly higher in exosomes from human bone marrow mesenchymal stem cells than in those from human pancreatic cancer cell line exosomes, and suggested that hsa-miR-143-3p might promote apoptosis and suppress cell growth and invasion in pancreatic cancer [29]. Ramanathan et al. detected that poor responders had lower levels of exosomal hsa-miR-338-5p and proposed exosomal hsa-miR-338-5p as a potential biomarker due to its stability and dysregulation in diseases, including complex regional pain syndrome, a chronic pain disorder with persistent inflammation [30].
The BPs associated with the target genes (mRNA) of DEexo-miRNA in blood and urine encompassed both positive and negative regulation of transcription from an RNA polymerase II promoter on a DNA template, as well as modifications to chromatin structure. The CCs involved included the nucleoplasm, cytosol, nucleus, cytoplasm, adhesive spots, and membranes. The MFs of these target genes were found to be diverse, involving protein binding (including kinase, domain-specific, serine/threonine, and tyrosine kinase interactions), ubiquitin protein ligase binding and transferase activity, as well as ATP, cadherin, GDP binding, and transcription factor activity, which includes sequence-specific DNA binding within the RNA binding transcriptional regulation region. The target mRNAs of DEexo-miRNAs in blood and urine play roles in various signaling pathways. These pathways include proteoglycans in cancer, axonal guidance, regulation of the actin cytoskeleton, cancer pathways, resistance to EGFR tyrosine kinase inhibitors, neurotrophic signaling pathways, endocytosis, attachment sites, cellular aging, autophagy in animals, and Salmonella infection.
Our analysis of blood and urine exosome sequencing revealed that the ncRNA content, including rRNA, tRNA, snoRNA, snRNA, miRNA, and other ncRNAs, was essentially identical. The database of eukaryotic DNA repeat sequences encompasses transposable elements, tandem repeat sequences (such as satellite sequences or microsatellites), scattered genomic repeat sequences, and some multicopy host genes (including rRNA, tRNA, histone genes, etc.). Additionally, the length distribution of candidate RNA (unique reads) exhibited slight variability. A total of 102 DEexo-miRNAs were identified between blood and urine, with 41 being more abundant in blood than in urine, and 61 showing higher levels in urine than in blood. The corresponding mRNAs of these miRNAs are involved in signaling pathways such as proteoglycans in cancer, axonal guidance, regulation of the actin cytoskeleton, and various cancer pathways. This study lays the groundwork for the identification of potential biomarkers derived from blood and urinary exosomes.